(43) Robust regression and outlier detection¶
In this example show how gmtregress can be used to determine a robust regression line using reweighted least squares and from this fit we are able to identify outliers with respect to the main trend. The result shows dinosaurs were large and dumb, humans and some monkeys pretty smart, and the rest of the mammals doing alright. Band shows 99% confidence internal on regression.
#!/usr/bin/env bash
# GMT EXAMPLE 43
#
# Purpose: Illustrate regression and outlier detection
# GMT modules: gmtregress, basemap, legend, text, plot
# Unix progs: grep, awk, sed
#
# set AWK to awk if undefined
AWK=${AWK:-awk}
# Data from Table 7 in Rousseeuw and Leroy, 1987.
gmt begin ex43
file=$(gmt which -Gc @bb_weights.txt)
gmt regress -Ey -Nw -i0:1+l $file > model.txt
gmt regress -Ey -Nw -i0:1+l $file -Fxmc -T-2/6/0.1 -C99 > rls_line.txt
gmt regress -Ey -N2 -i0:1+l $file -Fxm -T-2/6/2+n > ls_line.txt
grep -v '^>' model.txt > A.txt
grep -v '^#' $file > B.txt
$AWK '{if ($7 == 0) printf "%dp\n", NR}' A.txt > sed.txt
gmt makecpt -Clightred,green -T0/2/1 -F+c -N
gmt basemap -R0.01/1e6/0.1/1e5 -JX15cl -Ba1pf3 -Bx+l"Log@-10@- body weight (kg)" -By+l"Log@-10@- brain weight (g)" -BWSne+glightblue -Y10c
gmt plot -R-2/6/-1/5 -JX15c rls_line.txt -L+yt -Glightgoldenrod
sed -n -f sed.txt B.txt | gmt text -R0.01/1e6/0.1/1e5 -JX15cl -F+f12p+jRM -Dj10p
gmt plot -R-2/6/-1/5 -JX15c -L+d+p0.25p,- -Gcornsilk1 rls_line.txt
gmt plot rls_line.txt -W3p
gmt plot ls_line.txt -W1p,-
gmt plot -Sc0.4c -C -Wfaint -i0,1,6 model.txt
gmt text A.txt -F+f8p+jCM+r1 -B0
# Build legend
cat <<- EOF > legend.txt
H 18p,Times-Roman Index of Animals
D 1p
N 7 43 7 43
EOF
$AWK -F'\t' '{printf "L - C %d.\nL - L %s\n", NR, $NF}' B.txt >> legend.txt
gmt legend -DjBR+w6c+o0.4c -F+p1p+gwhite+s+c3p+r legend.txt --FONT_LABEL=8p
gmt basemap -R0.5/28.5/-10/4 -JX15c/5c -Y-7.5c -B+glightgoldenrod
gmt plot -Gcornsilk1 -W0.25p,- <<- EOF
>
0 -2.5
30 -2.5
30 2.5
0 2.5
> -Glightblue
0 -10
30 -10
30 -2.5
0 -2.5
EOF
$AWK '{print NR, $6, $7}' A.txt | gmt plot -Sb1q+b0 -W0.25p -C
gmt basemap -Bafg100 -Bx+l"Animal index number" -By+l"z-zcore" -BWSne
rm -f *.txt
gmt end show
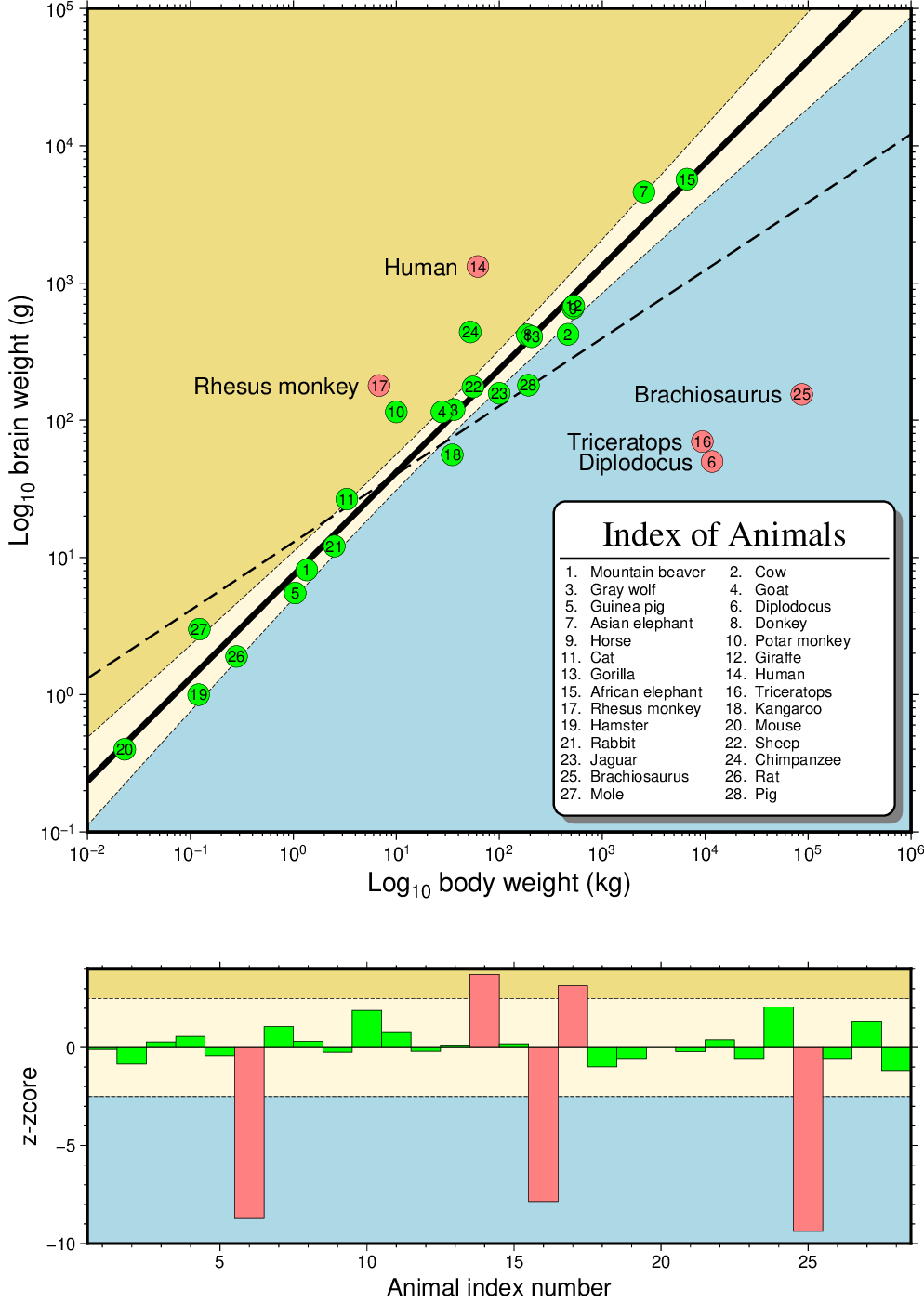
Robust Regression and Outlier Detection.¶